
MSU’s Frances Trail leads multi-funded investigation into evolution of infectious fungi
Fungi appeared on earth around 1.5 billion years ago. Today, there may be as many as 5 million fungal species, and they are found everywhere, from the mushroom that pops up
in your lawn to the food in the mold on the food in the back of your refrigerator.

A collaboration of international scientists and funding agencies, led in part by MSU’s College of Natural Science plant biologist Frances Trail, will investigate seven fungi whose common ancestor dates back 270 million years. While the spores of the ancestor germinated to grow in an ancient environment, each of the seven species branched out on their fungal family tree to make homes in very different places, from ants to plants to humans, sometimes causing disease in their hosts.
“We don’t understand the mechanisms of fungal adaptations to new hosts, thus our research is focused on identifying genes important to causing diseases in quite different organisms,” said Trail, professor in the Department of Plant, Soil and Microbial Sciences and associate chair in the Department of Plant Biology. “It was extraordinary to receive support for our study from so many agencies—the United States Department of Agriculture (USDA), the National Science Foundation (NSF) and the Binational Science Foundation (BSF).”
Trail will lead the 3-year, $730,000 USDA National Institute of Food and Agriculture (NIFA) grant that focuses on the disease-causing fungus Fusarium graminearum and a cool season grass fungal pathogen, Epichloë festucae. Carolyn Young, professor at the Noble Research Institute in Oklahoma, is co-investigator on the grant.
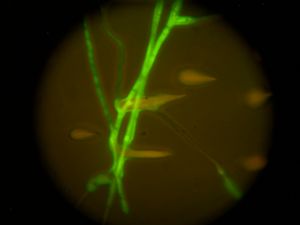
“These fungi evolved different morphologies that allow their spores to infect new hosts. These changes in their genomes helped them to occupy a new niche,” explained Trail, who has been working on fungal morphology and adaptations for over 30 years. “We want to understand the drivers of these changes by looking for key genes that have changed their expression and taken on new roles.”
The Trail Lab will focus on Fusarium graminearum, a fungus important to Michigan and the world because it infects wheat, barley and corn, causing $3 billion worth of crop damage in the United States between 1998 and 2003.
Two of the toxins produced by the Fusarium fungus, deoxynivalenol and zearalenone, are especially toxic to humans and animals.
“This fungus produces toxins that make the plant susceptible to infection by a spore germinating on the surface,” Trail said. “Once the fungus is inside the plant, it fills the grain with mycotoxins and causes illness if ingested by humans or animals. Growers and millers are concerned with eliminating these two mycotoxins from our food supply.”
Trail and her team will reconstruct evolutionary changes in morphology using comparative genomics, evolutionary biology and fungal pathogenesis. Their novel methodologies will lead them to the genes critical for differ entiated spore germination, growth and its ability to infect its host.
“Fungi share many genes that they have inherited from their ancestors, but as they evolve, some of these genes will take on new roles”, Trail said.

“We hypothesize that in many cases, new roles are accompanied by increases in expression in the lineage where these new roles make the fungus more pathogenic, so we will compare expression of common genes in lineages that arose from a common ancestor and look for those genes that increased in expression in just one lineage.”
“All of the fungi will be grown in a similar manner, so if one of them greatly increases the expression of a gene that they all have, then we would investigate the new role for that gene using gene knockout techniques ,” Trail explained. “That gene may used for something new, and maybe that new role is the adaptation that makes it pathogenic to a plant or an insect.”
Young will run parallel experiments with Epichloë, the fungus that causes “choking disease” in some cool season grasses. Seed transmitted Epichloë is known for causing fescue toxicosis in livestock grazing on tall fescue, while the choke forming species has negatively impacted orchardgrass seed production in Oregon.
“Epichloë will be a challenge as they don’t sporulate as readily as the other fungi in the study, and many that associate with plants are considered beneficial symbionts,” said Young, who has been working on this particular fungus for almost two decades. “But that’s also what makes them interesting to include. This grant provides a great opportunity to explore the biology of this symbiont while working alongside an excellent team of scientists.”
Other investigators include Jeffrey Townsend, Elihu Professor of Biostatistics at Yale, who will analyze Metarhizium anisopliae and Cordyceps militaris, the fungus responsible for zombie ants—a phenomenon effecting ant behavior and ultimately lethal. This work is funded by NSF. Oded Yarden, the Buck Family Chair of Plant Pathology at Hebrew University, Jerusalem, will use funds from the BSF to conduct investigations into the model fungi Neurospora crassa, the plant symbiont Trichoderma asperellum and Tolypocladium ophioglossoides, a pathogen of other fungal species.
For more information about the USDA’s NIFA AFRI grant, please visit: https://nifa.usda.gov/program/agriculture-and-food-research-initiative-afri.
View original article
